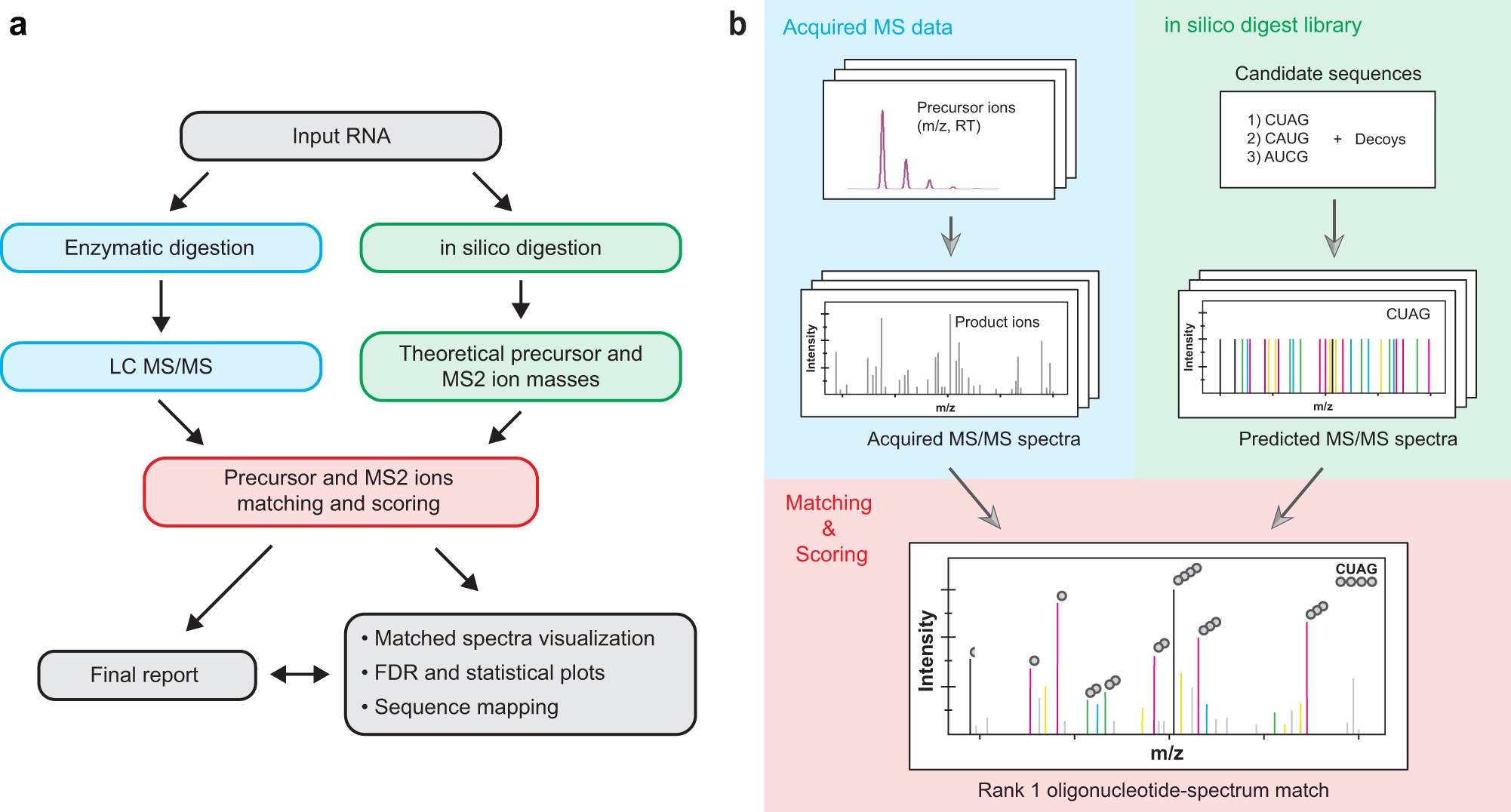
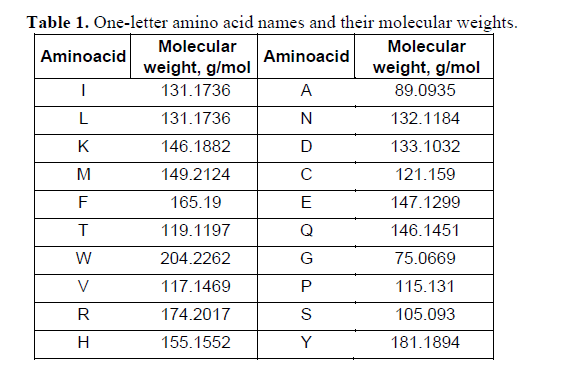
ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis | PLOS Computational Biology
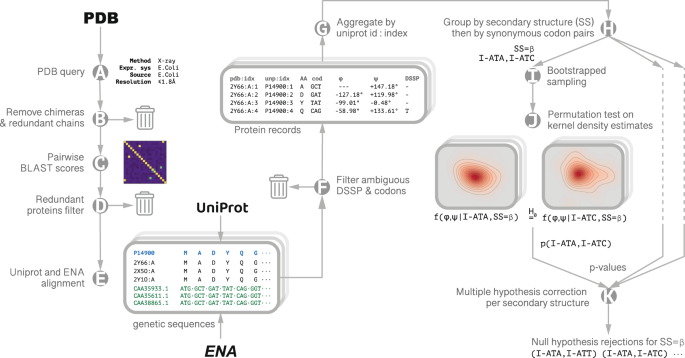
Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon | Nature Communications
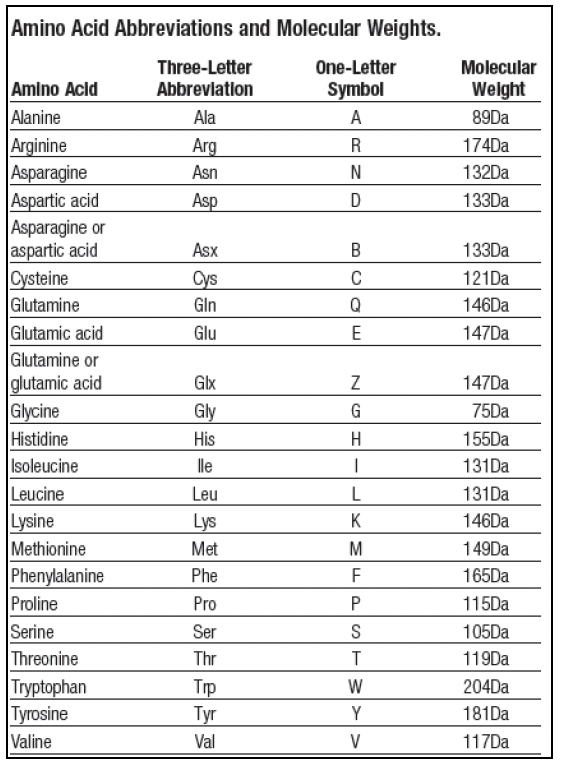
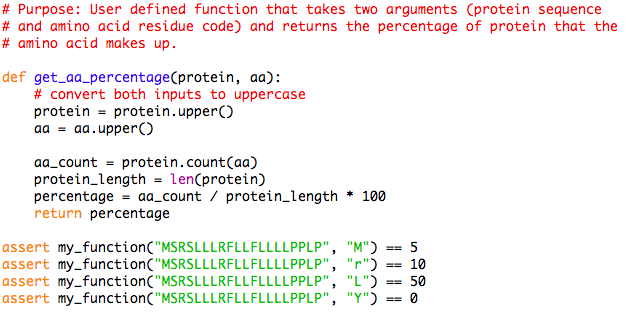
GitHub - efr-essakhan/pepfeature: A Python package that has functions for featue calculation of protein sequences. Features that are calculated by this package are niched for Epitope prediction.

Molecules | Free Full-Text | PepFun: Open Source Protocols for Peptide-Related Computational Analysis
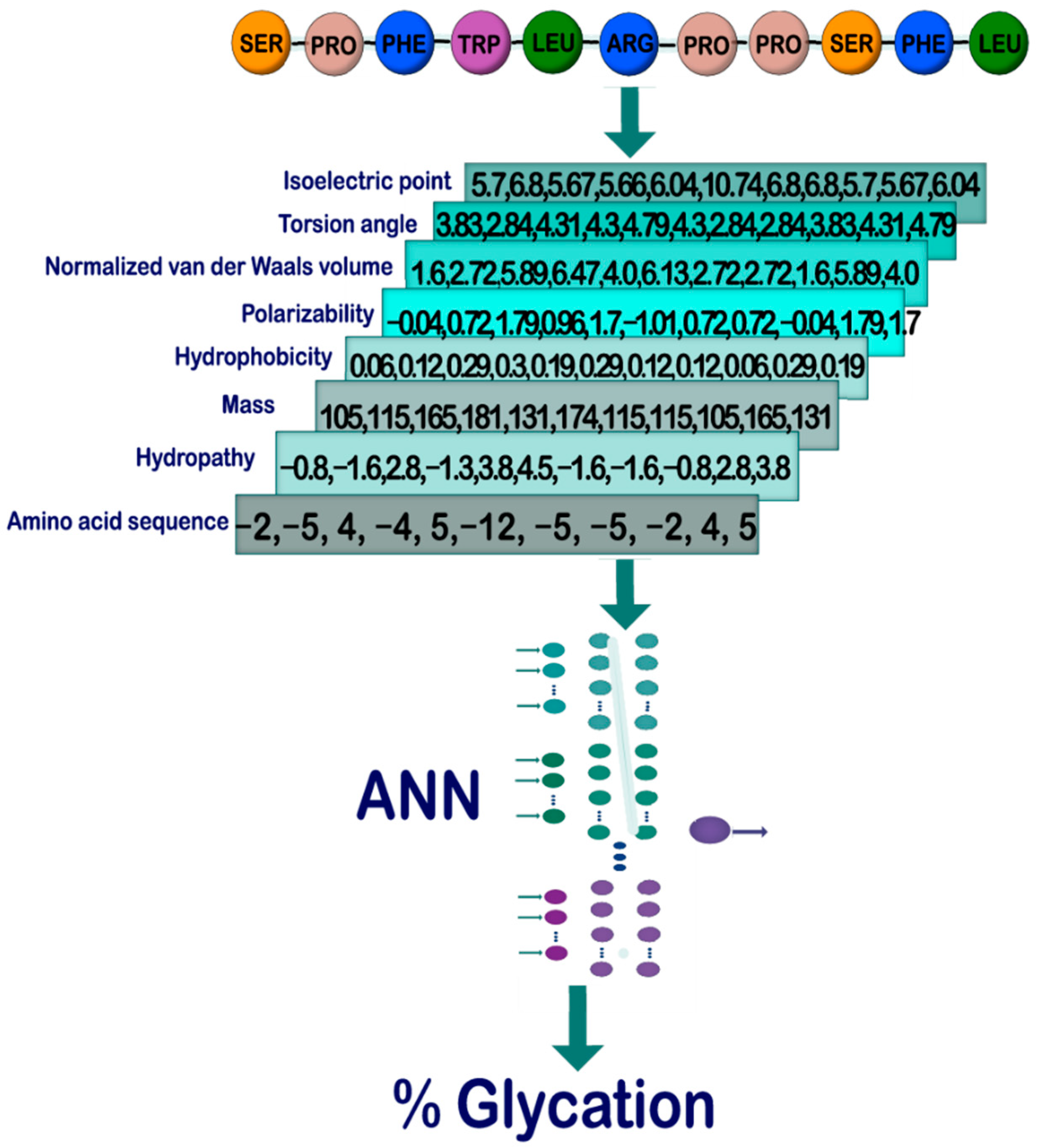
Sensors | Free Full-Text | On the Prediction of In Vitro Arginine Glycation of Short Peptides Using Artificial Neural Networks

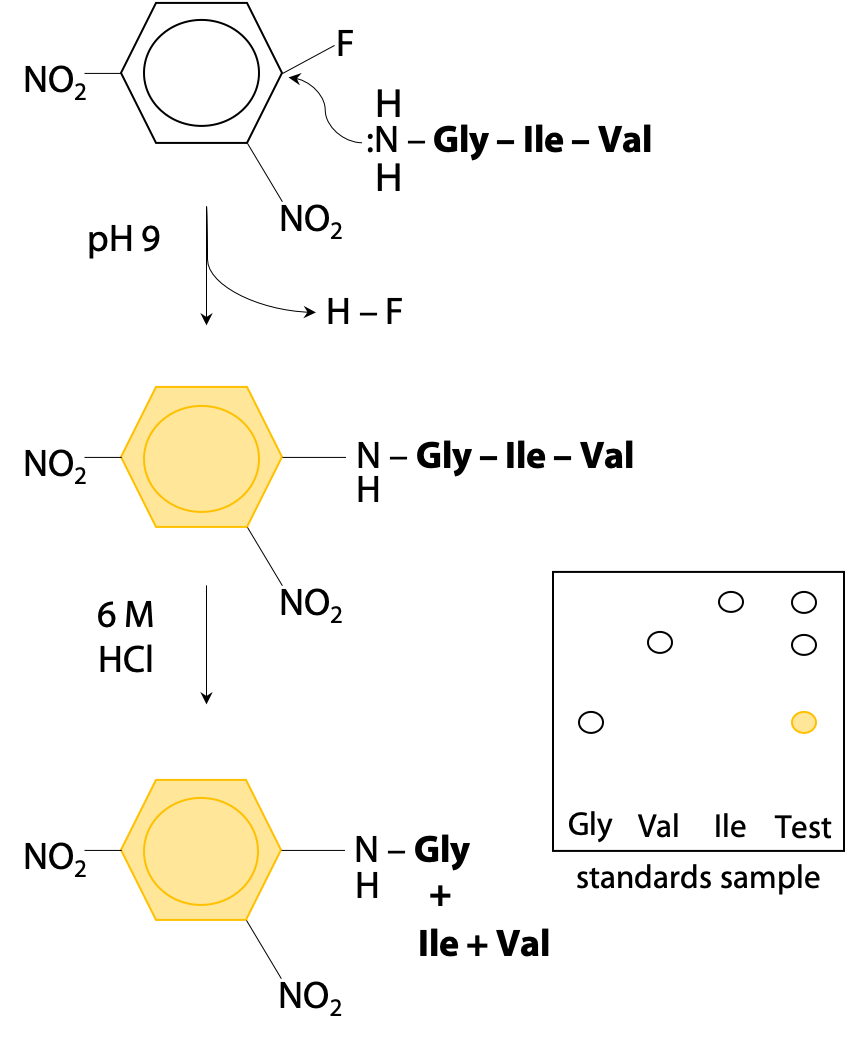
Frontiers | Essentially Leading Antibody Production: An Investigation of Amino Acids, Myeloma, and Natural V-Region Signal Peptides in Producing Pertuzumab and Trastuzumab Variants

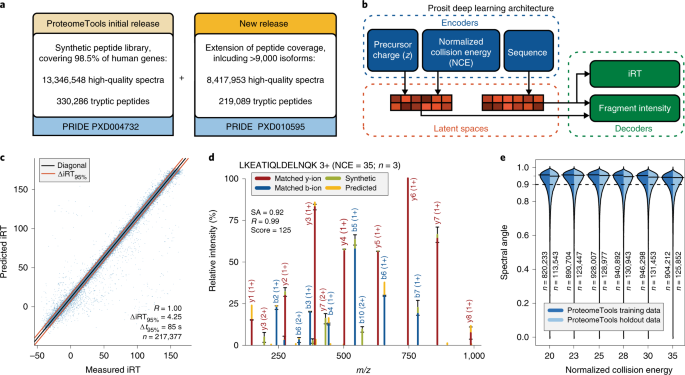
Data acquisition and database search. For a given experimental MS/MS... | Download Scientific Diagram